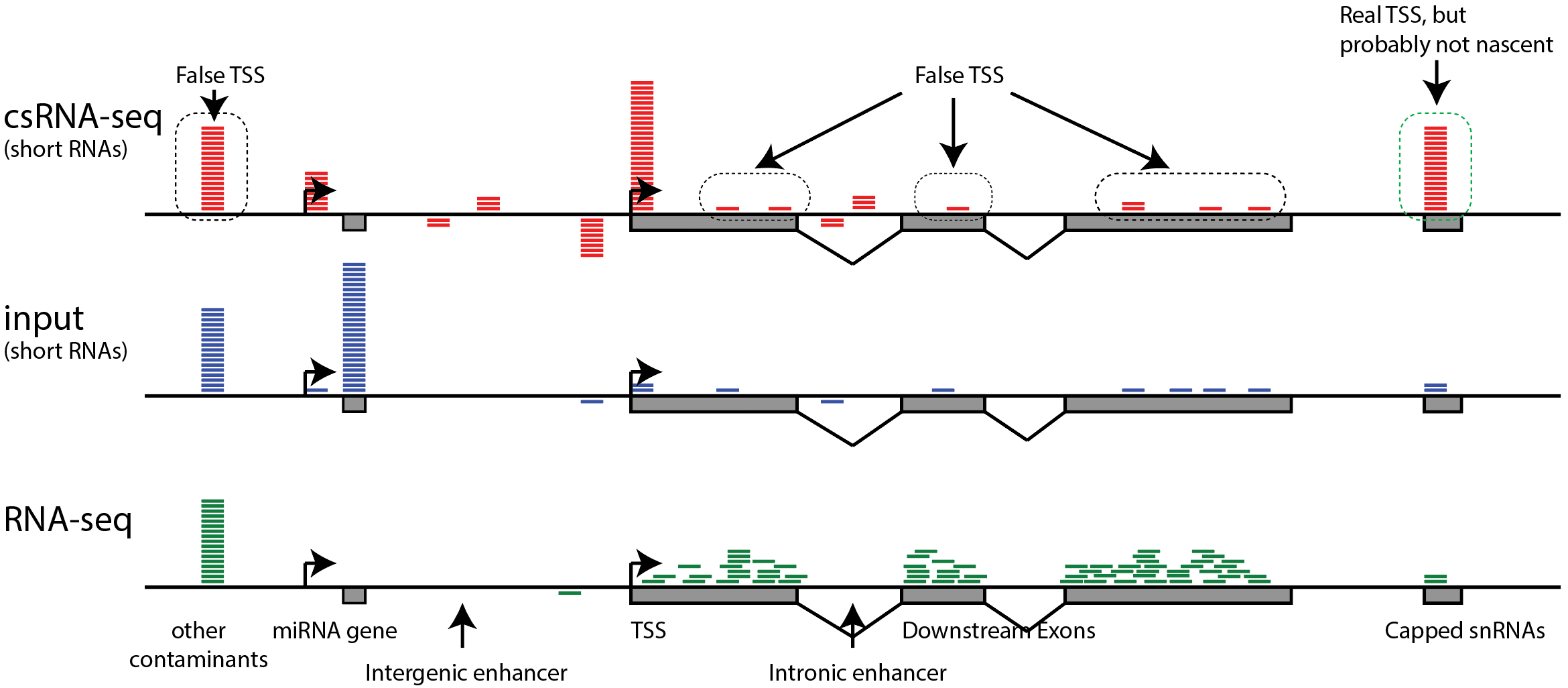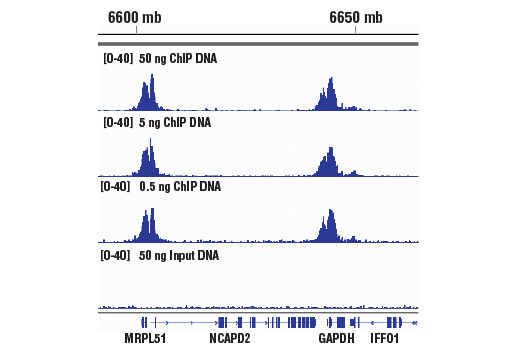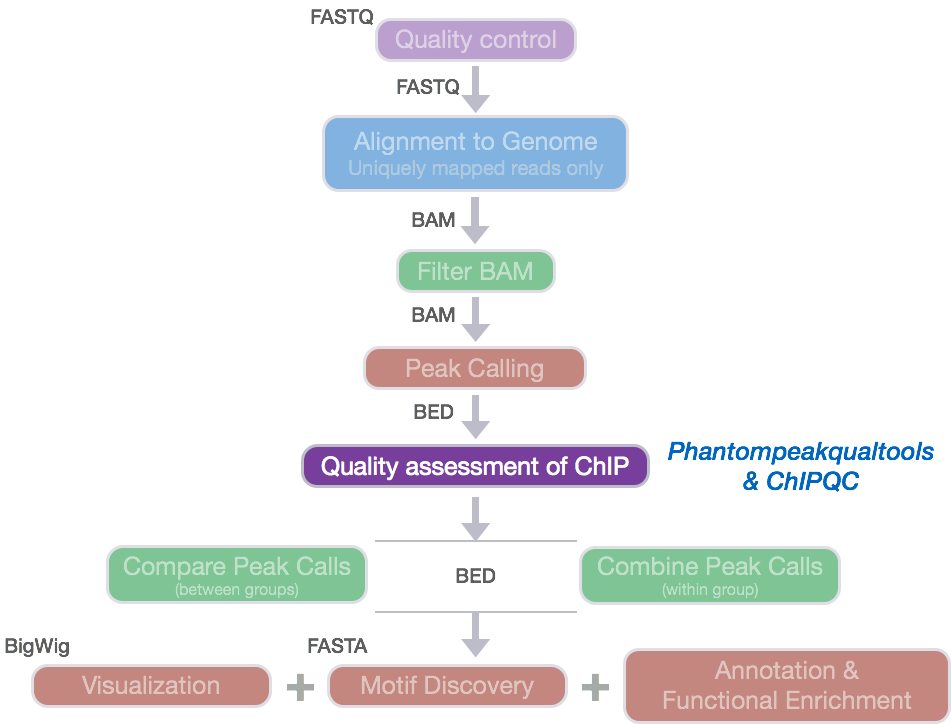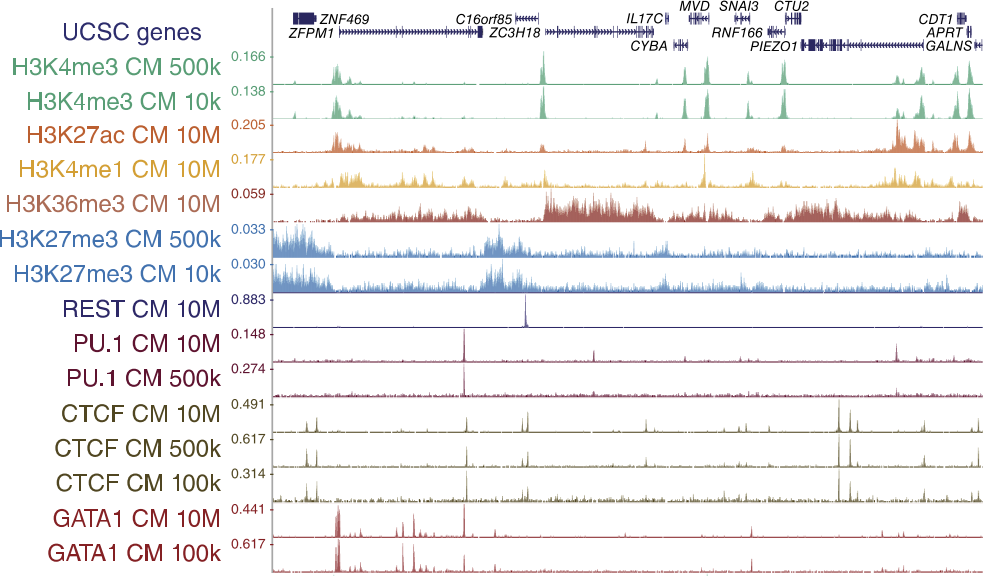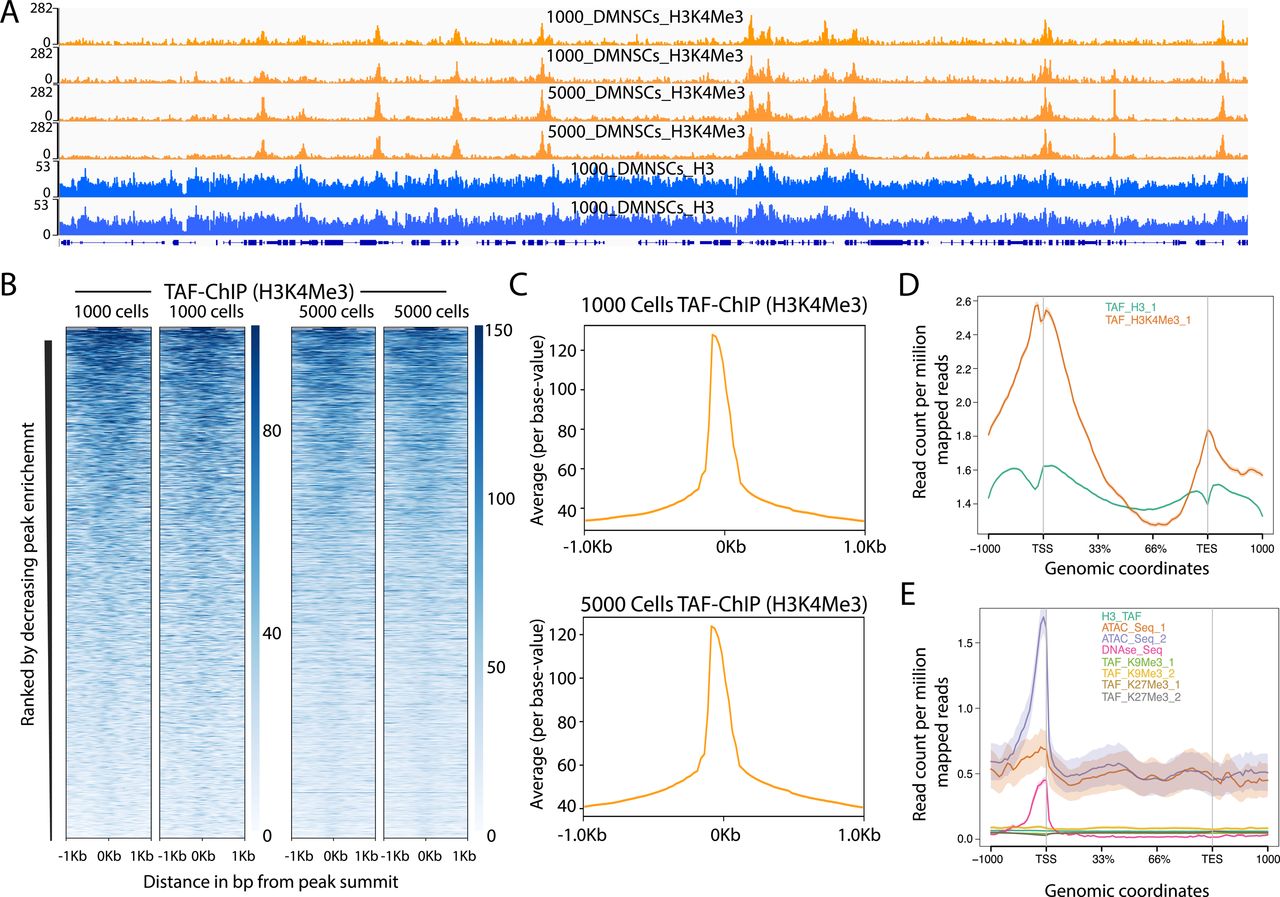
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance
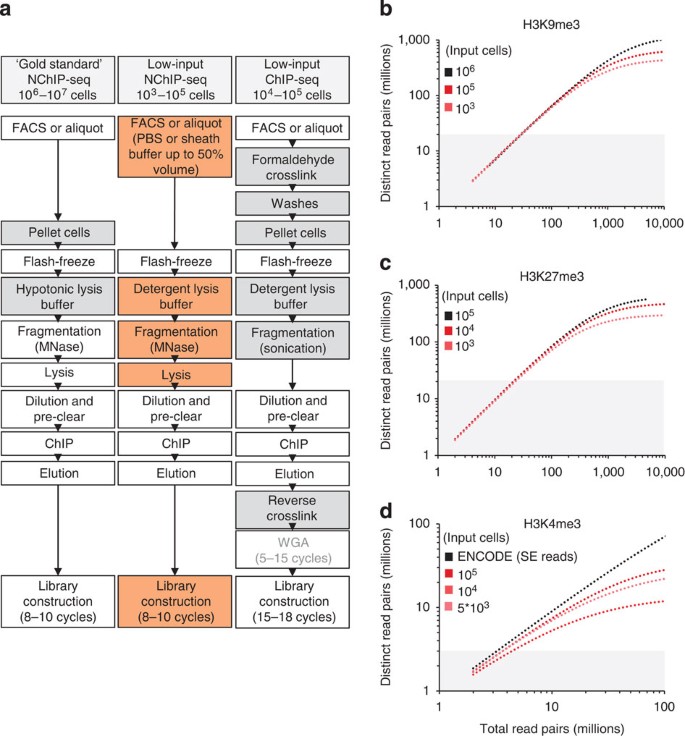
An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations | Nature Communications

ChIP-seq protocol for sperm cells and embryos to assess environmental impacts and epigenetic inheritance - ScienceDirect

ChIP-seq profiles of AGO2 in S2 and S3 cells at BX-C. AGO2 ChIP-seq... | Download Scientific Diagram
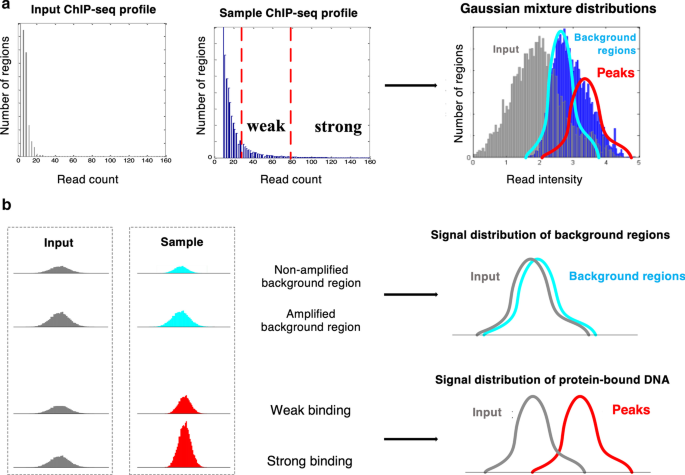
ChIP-BIT2: a software tool to detect weak binding events using a Bayesian integration approach | BMC Bioinformatics | Full Text

TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance